Pharmacokinetic/Pharmacodynamic Modeling and Systems Analysis
David Z. D’Argenio, Ph.D.
Project Leader
Systems modeling provides a quantitative framework for understanding the molecular and cellular mechanisms that underlie how medicines produce their effects (quantitative systems pharmacology – QSP) and for characterizing the interpatient variability observed in response to treatment (pharmacometrics). The overall goal of Core Project #1 is to develop, evaluate, apply, and disseminate methods of QSP and pharmacometrics to advance model-based discovery, development and precision use of medicines, leading to more effective treatments for life threatening diseases. The methods for QSP developed and applied in this project are aimed at quantifying the mechanisms of action of different therapeutic modalities including small molecule and proteins drugs, cell and gene therapies, and protein degraders. Our advances in pharmacometrics are applied to better understand the absorption, distribution, metabolism, and elimination of therapeutic agents (pharmacokinetics), how medicines produce their effects (pharmacodynamics), and to quantify the contribution of genetic and epigenetic factors in determining the therapeutic response to treatment in individual patients and populations. The advanced systems modeling and associated computational methods we have developed are tailored for the challenging spare data environment of model-based discovery, development and therapeutics. Application areas include oncology, as well as infectious, autoimmune and metabolic diseases. Several current projects are outlined below.
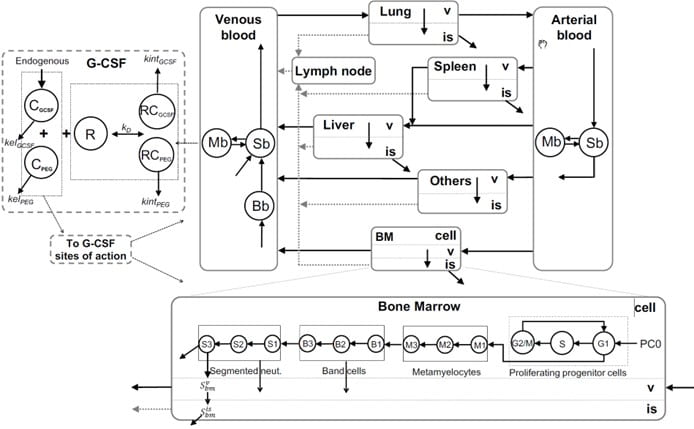
Physiological Modeling of Granulopoiesis to Predict Clinical Drug Induced Neutropenia from in vitro Bone Marrow Studies
Many anticancer therapies, including molecularly targeted therapies, disrupt neutrophil homeostasis through their myelosuppressive actions, resulting in neutropenia. We have developed a whole-body model of granulopoiesis and its regulation that incorporates the circulatory disposition of neutrophils together with organ-specific transport in liver, spleen, lung and bone marrow. The bone marrow subsystem includes a cell cycle model of progenitor cells, which allows for a mechanistic representation of the action of relevant anticancer drugs derived from in vitro bone marrow toxicity studies, thereby providing a model-based framework for predicting drug induced neutropenia in humans from preclinical studies.
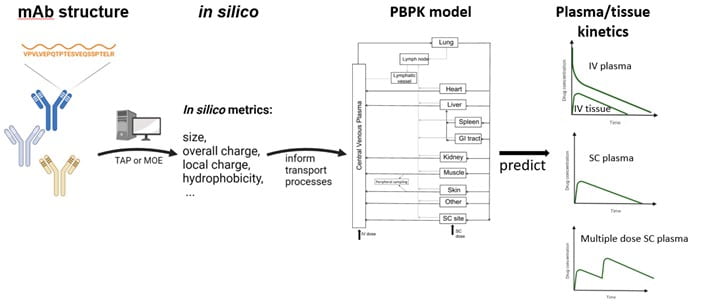
Predicting Monoclonal Antibody Pharmacokinetics in Humans from in vitro Physiochemical Properties
Use of the subcutaneous route for administering monoclonal antibodies to treat chronic conditions has been hindered due to our incomplete understanding of the fundamental mechanisms controlling mAb absorption from the SC site, and the limited translatability of preclinical studies. We have developed an integrated physiologically based modeling framework for predicting pharmacokinetics and bioavailability of subcutaneously administered monoclonal antibodies based on in silico structure-derived metrics characterizing antibody size, overall charge, local charge and hydrophobicity.
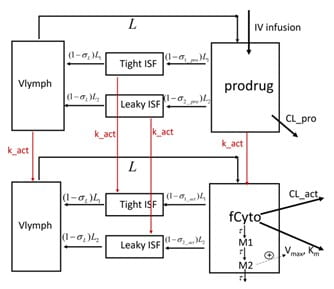
Pharmacokinetics of the Engineered Fc-IL-15 Proteolytically Activated Cytokine Prodrug
Stimulatory cytokine-based therapeutics can activate lymphocytes, including natural killer cells and CD8+ T cells in the immunosuppressive tumor microenvironment (TME), resulting in higher cancer immunotherapy response rates. Their clinical use, however, has been hindered by unfavorable pharmacokinetics due to the “cytokine sink”, systemic toxicity, and a narrow therapeutic index. To enhance TME specificity and extend their half-life, cytokine prodrugs have been designed wherein the binding of the cytokine to lymphocytes is masked by a masking moiety, which can be cleaved by extracellular proteases in the TME [3]. A mechanistic model was developed to describe the PK of an engineered an Fc-IL-15 proteolytically activated cytokine prodrug and its active cytokine in cynomolgus monkeys.
ADAPT Software Platform for PK/PD Systems Modeling and Analysis
An overarching goal of Core Project #1 is to provide advanced modeling and analysis methods to the broader biomedical research community through the ADAPT Software for PK/PD Systems Modeling and Analysis, thereby enhancing the basic and clinical research efforts of other investigators – the raison d’être of a Research Resource. Toward this end, we continue to develop, disseminate and support the ADAPT software for PK/PD systems modeling and analysis. The ADAPT software platform includes computational tools for dynamic systems modeling in QSP and pharmacometrics, and contains applications for individual and population model simulation, likelihood and Bayesian model estimation, experiment design, and hierarchical nonlinear modeling analysis of clinical trial data. ADAPT is used by researchers and clinical scientists in universities, industry and government agencies to advance the discovery, development and precision use of medicines.
Selected Recent Publications
Li R, Craig M, D’Argenio DZ, Betts A, Mager DE, Maurer TS. Editorial: Model-informed Decision Making in the Preclinical Stages of Pharmaceutical Research and Development. Front Pharmacol. 14:1184914. 2023.
N. Schweighofer; D. Lyn; H. Luo; D.Z. D’Argenio; C. Winstein. Long-Term Forecasting of a Motor Outcome Following Rehabilitation in Chronic Stroke via a Hierarchical Bayesian Model of Motor Learning. Journal of NeuroEngineering and Rehabilitation. 20(1):83, 2023. doi: 10.1186/s12984-023-01202-y.
S. Hu, A. Datta-Mannan and D. Z. D’Argenio. Monoclonal Antibody Pharmacokinetics in Cynomolgus Monkeys Following Subcutaneous Administration: Physiologically Based Model Predictions from Physiochemical Properties. AAPS Journal. 25(1):5, 2022. doi: 10.1208/s12248-022-00772-4.
M. Weiss; D.Z. D’Argenio, W. Siegmund. Analysis of Complex Absorption after Multiple Dosing: Application to the Interaction Between the P-glycoprotein Substrate Talinolol and Rifampicin. Pharmaceutical Research. 39:3292-330, 2022. doi.org/10.1007/s11095-022-03397-6
S. Hu, A. Datta-Mannan and D.Z. D’Argenio. Physiologically-Based Modeling to Predict Monoclonal Antibody Pharmacokinetics in Humans from in vitro Physiochemical Properties. mAbs. 14:1, 2056944, 2022. doi: 10.1080/19420862.2022.2056944.
W. Chen, B. Boras, T. Sung, W. Hu, M.E Spilker, and D.Z. D’Argenio. A whole-body circulatory neutrophil model with application to predicting clinical neutropenia from in vitro studies. CPT Pharmacometrics Syst. Pharmacol. 00:1-11, 2021. doi:10.1002/psp4.12620.
N.M. Smith, A. Chan, L.A. Wilkinson, H.C. Chua, T.D. Nguyen, H. de Souza, A.P. Shah, D.Z. D’Argenio, K.A. Mergenhagen. Open-Source Maximum a Posteriori-Bayesian Dosing AdDS to Current Therapeutic Drug Monitoring: Adapting to the era of Individualized Therapy. Pharmacotherapy. 11:953-963, 2021. doi: 10.1002/phar.2631.
Y.O. Kim, K-J Kim, D.Z. D’Argenio and E.D. Crandall.. Characteristics of Passive Solute Transport Across Primary Rat Alveolar Epithelial Cell Monolayers. Membranes. 11: 331, 2021. doi.org/10.3390/membranes11050331
S. Hu, Y. Lu, A. Tura, G. Pacini and D.Z. D’Argenio. An Analysis of Glucose Effectiveness in Subjects with or without Type 2 Diabetes via Hierarchical Modeling. Frontiers in Endocrinology. 12:641713, 2021. doi: 10.3389/fendo.2021.641713.
W. Chen, D.Z. D’Argenio, A. Sipos, K-J Kim, and E.D. Crandall. Biokinetic Modeling of Polystyrene Nanoparticle Interactions with Primary Rat Lung Alveolar Epithelial Cell Monolayers: Uptake from Apical Fluid, Intracellular Processing and Egress from Cells. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 330:R36-R43, 2021. doi: 10.1152/ajpregu.00184.2020.
A G. Drusano, S. Kim, M. Almoslem, S. Schmidt, D.Z. D’Argenio, J. Myrick, B. Duncanson, J. Nole, D. Brown, C. Peloquin, M. Neely, W. Yamada, and A. Louie. The Funnel: A screening technique for identifying optimal two-drug combination chemotherapy regimens. Antimicrobial Agents and Chemotherapy. 65(2):e02172-20, 2021. doi: 10.1128/AAC.02172-20.
W. Chen, B. Boras, T. Sung, W. Hu, M.E Spilker, and D.Z. D’Argenio. Predicting chemotherapy-induced neutropenia and granulocyte-colony stimulating factor response using model-based in vitro to clinical translation. The AAPS Journal. 22:143, 2020. doi: 10.1208/s12248-020-00529-x.
S. Jaisue, C. Pongsakul, D.Z. D’Argenio and P. Sermsappasuk. Population pharmacokinetic modeling of vancomycin in Thai patients with heterogenous and unstable renal function. Therapeutic Drug Monitoring. 42:856-865, 2020. doi: 10.1097/FTD.0000000000000801.
C. Wang, C. Winstein, D.Z. D’Argenio and N. Schweighofer. The efficiency, efficacy, and retention of motor training in chronic stroke neurorehabilitation and neural repair. Neurorehabilitation & Neural Repair. 34:881-890, 2020. doi: 10.1177/1545968320948609.
S. Hu and D.Z. D’Argenio. Predicting monoclonal antibody pharmacokinetics following subcutaneous administration via whole-body physiologically-based modeling. Journal of Pharmacokinetics and Pharmacodynamics. 47:385-409, 2020. doi:10.1007/s10928-020-09691-3.
W. Chen, B. Boras, T. Sung, Y. Yu, J. Zheng, D. Wang, W. Hu, M.E. Spilker, D.Z. D’Argenio. A Physiological Model of Granulopoiesis to Predict Clinical Drug Induced Neutropenia from in vitro Bone Marrow Studies. Journal of Pharmacokinetics and Pharmacodynamics. 27:163-182, 2020. doi: 10.1007/s10928-020-09680-6.